- 3-5 Impasse Reille 75014 Paris France
- info@depixus.com
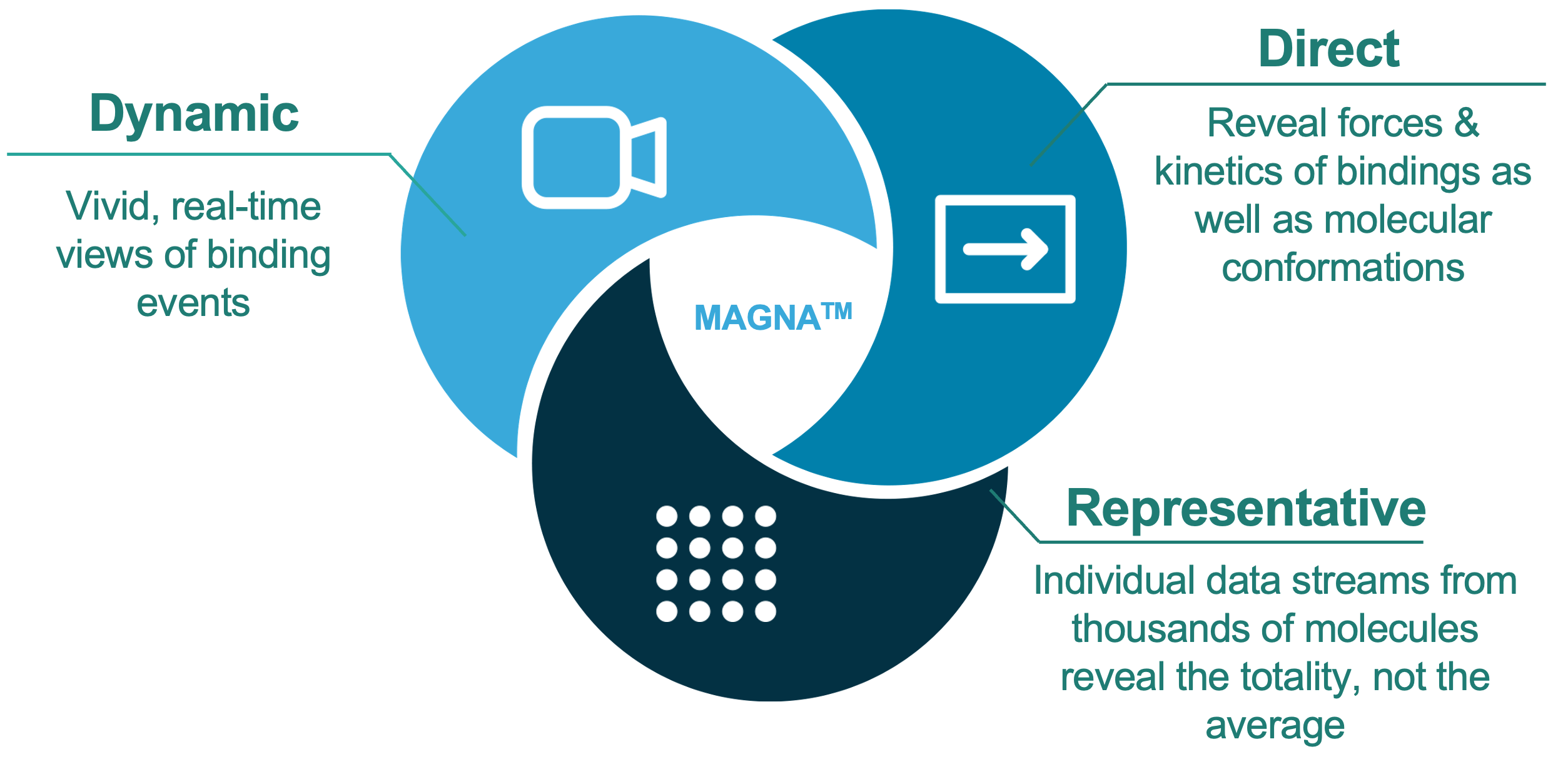
MAGNA™, the first technology to deliver direct, real-time measurement of dynamic interactions across thousands of individual molecules.

See our highly versatile MAGNA™ technology ‘in action’ in this short video :
MAGNA™ comprises a benchtop analytical instrument, flow-cell consumables and a data handling system. Analyses can be configured in three distinct assay modes, depending on the requirements of the experiment:
This mode is used to measure the strength on binding between two molecules.
Pairs of molecules to be tested are tethered at a fixed distance apart along oligonucleotide strands attached to the flow-cell floor at one end and bound to paramagnetic beads at the other. When no magnetic force is applied, the oligo is relaxed so that the tethered molecules are able to interact. As the magnetic force is applied, they are pulled apart and their binding force is measured.
Applications include: Protein-protein, protein-antibody, protein-small molecule interactions


Structure mode is used to test whether a target’s 3D structure is stabilized or destabilized by a binding partner such as a protein or small molecule.
In this mode, native target molecules of interest are bound at one end to the floor of a flow-cell with short oligonucleotides and to paramagnetic beads at the other. Interacting molecules are then added and the molecules are unfolded using a fixed magnet above the flow-cell, with the position of each magnetic bead tracked by an optical system.
Applications include: RNA-targeted drug discovery and RNA binding protein research.
Binding position mode is used to determine the precise location on an RNA or DNA target that binds one or more interacting molecules.
Strands of DNA or RNA of interest are prepared with a hairpin at one end and a Y-shaped handle at the other. One branch of the Y-shape is attached to the floor of the flow-cell and the other to a paramagnetic bead. As the magnetic force is applied, the hairpins unzip and can be interrogated with interacting molecules, such as antibodies that bind to specific base modifications, or short oligos that can act as bar codes to determine the target’s sequence. The target molecules refold as the force is released, pausing when a binder is encountered. The positions of these pauses are measured to single base precision, enabling the exact location of the interactions to be determined.
Applications include: Nucleic acid QC and DNA-binding proteins
